Seed ORDER
Seed - Oilseed Rape Developmental Expression Resource
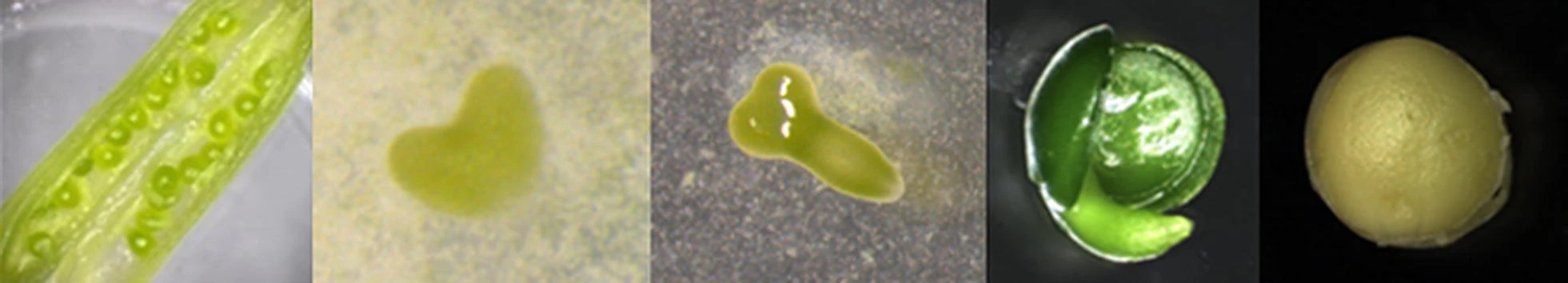
The Seed - Oilseed Rape Developmental Expression Resource (Seed ORDER) was created to provide access to the developmental transcriptome of Brassica during seed maturation.
Guide
The "About" tab presents information about how the experiment was carried out, such as which Brassica varieties were grown, how the samples were collected and how gene expression levels were quantified.
The "How To Use" tab explains how to use the website to search the database. This is the place to go if you have questions about the website's user interface.
The final three tabs, "Search", "BLAST Search" and "Table", are the tabs you'll use to search the database.
Contact
Please could you direct enquiries to Rachel Wells and Smita Kurup.
AionPlot
Seed ORDER is an instance of the time series visualization and dissemination platform AionPlot.
About
This web app facilitates the interrogation of time series data collected from Brassica napus and Brassica oleracea.
The data was collected from the B. napus cultivars, Express 617 (a modern winter cultivar), Zhongshuang 11 (ZS11) (a semi-winter cultivar), Ragged Jack (a heritage kale cultivar), Stellar DH (a spring cultivar), together with data from the B. oleracea cultivar DH1012 (rapid-cycling double haploid line from a cross between B. oleracea Alboglabra ‘A12’and B. oleracea Italica ‘Green Duke’), with the aim of deciphering how the seed development gene regulatory network is constructed, and how it differs from the network elucidated in Arabidopsis thaliana.
An overview of the plant growth regime and data processing pipeline is provided below.
Growth Details
Plants were germinated and grown under controlled environmental conditions with a 16h photoperiod at 18°C/15°C day/night temperatures, respectively, under 400 W HQI lighting (225 µmol m-2s-1). The relative humidity was maintained at 70%. At 3 weeks old, the plants from the winter and semi-winter cultivars were transferred to vernalization. After three weeks (ZS11), six weeks (Express 617), or 12 weeks (Ragged Jack) weeks of vernalization, plants were returned to the controlled environment rooms until maturity.
Sampling Details
Tissue collections, manual pollinations and dissections to obtain pre-fertilization floral buds, ovules and ovary walls, as well as heart, torpedo, green and mature stage embryos, endosperms and seed coats were followed as described in Siles et al. (2020). Leaves and floral apex at 21 days after sowing, as well as valve walls at "embryo green stage" were collected from each Brassica spp genotype. The silique valves in green stage, that corresponds to when the embryos are in green developmental stage, were also collected. For each time point and tissue types, three biological replicates were performed. At each time point, relevant tissue was extracted using sterile forceps over a clean, sterile tile placed on a bed of ice within a 14 cm diameter plastic petri dish to ensure the samples were cold throughout the dissection procedure. The samples were immediately frozen in liquid nitrogen and stored at -80°C.
RNA Extraction Details
High quality total RNA was extracted as described in Siles et al. (2020). All the collected valve tissue for each replicate was pooled together and finely powdered by grinding in liquid nitrogen in an autoclaved mortar and pestle. TRIzol reagent (ThermoFisher Scientific, Waltham, USA) was used for the extraction of valve in green stage RNA as described in Siles et al. (2020) for green stage seed coat. The RNA was subsequently treated for DNAse using the TURBO DNA-free kit (ThermoFisher Scientific, Waltham, USA). RNA integrity number (RIN) was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) at Rothamsted Research.
Total RNA samples were processed by Novogene (Beijing, China). Libraries were sequenced on the Illumina HiSeqX, resulting in 150bp paired end reads. Sequencing data is available from the Sequence Read Archive with accession number PRJNA1083450.
Expression Quantification
Quality control analyses were performed for raw RNA-Seq reads. The raw expression reads were then analysed using an RNA-seq alignment pipeline consisting of Trimmomatic (v0.39), hisat2 (v2.1.0) and stringtie (v2.1.1). "Trimmomatic" trims adapter sequences and quality checks RNA-seq sequencing reads, "hisat2" is a splice-aware aligner, which is able to align sequencing reads that span a splice site to genomic reference sequences, and "stringtie" quantifies the gene expression from the aligned sequencing reads for each transcript.
The expression data shown here is the mean TPM across replicates for each tissue/stage with the error bars showing the standard error of the mean of the pertinent TPM values.
Mappings to published gene models
The Brassica napus and Brassica oleracea cultivars were aligned to Darmor-bzh v4.1 and TO1000, respectively. Both of these genome references are available from Ensembl Plants, which also provided the homology data between the genotypes.
The published gene models are:
How To Use
In order to search the database, please use the "Search", "BLAST Search" and "Table" tabs.
Search
The "Search" tab allows users to plot the expression of Brassica genes across developmental time. To search for Brassica genes, type the name or abbreviation of a gene of interest and select the gene from the dropdown list. Although you may search for Brassica genes directly, ortholog and paralog mappings for the genome references are also available. The included gene model mappings are listed on the "About" tab.
You may enter multiple genes at once. Brassica genes which show homology to your genes of interest will be displayed below the search box. To plot the expression of a Brassica gene, click on its name. Hovering over the button or the name of the gene in the plot legend will show the Brassica gene's chromosome.
Gene names can be entered when searching, e.g., PDI3 will find the Arabidopsis gene, AT1G52260, which upon clicking will populate the bottom right of the screen with the Brassica orthologs. The colour of the button indicates the homology status of the Brassica gene. Brassica genes that Ensembl Plants considers to be of high confidence are coloured green with lower confidence orthologs coloured blue, e.g.,
and
These button colours are intended as a guide to aid users. The data is a subset of the homology data downloaded from Ensembl Plants and stated in the "About" tab. From this data, we also retained certain B. napus and B. oleracea paralogs, which will be displayed when searching for a Brassica gene and are displayed without highlighting, e.g.,
After selecting one or more genes, choose from the options in the drop-down lists. These choices will update the way in which the graphs are plotted, e.g., which data are shown in the rows. To focus on a single cultivar, select "Tissue" from the first drop-down, "None" from the second, and then choose the desired cultivar from the one below it.
Clicking the "Display error bars?" check-box will show/hide the error bars, which shows the standard error of the mean of the TPM values. To focus on specific tissues the sliders can be moved to hide certain ones. Finally the plot, its data and sequence FASTA can be downloaded by clicking the appropriate button.
The tissue and stage are encoded using a number from 1 to 8. The correspondence is as follows:
| ID | Tissue | Stage |
|---|---|---|
| 1 | Apex (21 days) | Pre-fertilisation |
| 2 | Leaf (21 days) | Pre-fertilisation |
| 3 | Gynoecia wall (24h before anthesis) | Pre-fertilisation |
| 4 | Ovules (24h before anthesis) | Pre-fertilisation |
| 5 | Embryo | Heart |
| 5 | Seed coat | Heart |
| 5 | Endosperm | Heart |
| 6 | Embryo | Torpedo |
| 6 | Seed coat | Torpedo |
| 6 | Endosperm | Torpedo |
| 7 | Embryo | Green |
| 7 | Seed coat | Green |
| 7 | Endosperm | Green |
| 7 | Silique wall | Green |
| 8 | Embryo | Mature |
| 8 | Seed coat | Mature |
BLAST Search
DNA sequences may be used to query the database. To do this, copy and paste your sequence of interest into the text box. The information box will indicate the number of Brassica genes that show homology to your sequence. To plot the expression of these genes, navigate back to the "Search" tab. The genes that show homology to the sequence you entered in the "BLAST Search" tab will be displayed as "BLAST Hits".
Table
Displays a table of details about the Brassica genes currently selected in the "Search" tab. Clicking the symbol will expand the row to reveal further homology information about that gene. The row colouring matches the button colouring detailed above in the section explaining the "Search" tab. Homologs between Brassica species are shown using the same colour scheme. Paralogs within the Brassica species are also shown and are not colour-coded. However, only paralogs from the Ensembl Plants homology data where the Brassica gene is orthologous to an Arabidopsis gene or where the average identity is 65% or more are included.
The columns, Gene Order Conservation (GOC) score, Whole Genome Alignment (WGA) coverage, Query identity and Target identity are from the Ensembl homology data and are used by Ensembl to QC the orthologs. How the values are calculated and used is explained in detail in the Orthology quality-controls help page.
Search
BLAST Search
Please enter a sequence in the box above to search the database with BLAST.